install.packages(
c(
"arrow",
"bench",
"devtools",
"dplyr",
"DT",
"duckdb",
"fs",
"knitr",
"rmarkdown")
)
devtools::install_github("Infectious-Disease-Modeling-Hubs/hubUtils")Hub model output re-partition benchmarks
Summary
- Query makes a big difference in how partitioning performs (which makes sense)
- Just converting to Parquet has major performance benefits, often more than re-partitioning.
- Perfomance of duckDB dependes on how much of the data is being queried. For small queries, it is faster than parquet arrow datasets, but for larger queries, it is slower.
- Csv has the worst perfomance.
Load packages
Install benchmarking dependencies
library(hubUtils)
library(dplyr)
library(duckdb)Connect to test hubs
Define paths to test hubs
Baseline std hubverse hub
baseline_hub_path <- "repartioned_hubs/flusight-hub_std_csv"Baseline parquet hubs
baseline_parquet_hub_path <- "repartioned_hubs/flusight-hub_model_id-reference_date_parquet/model-output"Reference Date re-partitioned hubs
Will be connecting using connect_model_output function as connect_hub needs some additional work to correctly connect to re-partitioned hubs.
Hence the paths to the repartitioned hubs point to the model-output directory of each hub.
ref_date_csv_hubpath <- "repartioned_hubs/flusight-hub_reference_date_csv/model-output"
ref_date_parquet_hubpath <- "repartioned_hubs/flusight-hub_reference_date_parquet/model-output"Target re-partitioned hubs
target_csv_hubpath <- "repartioned_hubs/flusight-hub_target_csv/model-output"
target_parquet_hubpath <- "repartioned_hubs/flusight-hub_target_parquet/model-output"DuckDB database path
duckdb_hub_path <- fs::path("repartioned_hubs", "flusight-hub.duckdb")Connect to hubs
Baseline hub
baseline_csv_hub_con <- connect_hub(baseline_hub_path)
baseline_csv_hub_con── <hub_connection/FileSystemDataset> ──• hub_name: "US CDC FluSight"• hub_path: 'repartioned_hubs/flusight-hub_std_csv'• file_format: "csv(613/613)"• file_system: "LocalFileSystem"• model_output_dir: "repartioned_hubs/flusight-hub_std_csv/model-output"• config_admin: 'hub-config/admin.json'• config_tasks: 'hub-config/tasks.json'── Connection schema hub_connection with 613 csv files
reference_date: date32[day]
target: string
horizon: int32
target_end_date: date32[day]
location: string
output_type: string
output_type_id: string
value: double
model_id: stringTotal number of rows in hub:
baseline_csv_hub_con |>
collect() |>
nrow()[1] 3355802baseline_parquet_hub_con <- arrow::open_dataset(baseline_parquet_hub_path)
baseline_parquet_hub_conFileSystemDataset with 613 Parquet files
target: string
horizon: int32
target_end_date: date32[day]
location: string
output_type: string
output_type_id: string
value: double
model_id: string
reference_date: stringReference Date hubs
ref_date_csv_con <- arrow::open_dataset(ref_date_csv_hubpath,
format = "csv"
)
ref_date_csv_conFileSystemDataset with 19 csv files
target: string
horizon: int64
target_end_date: date32[day]
location: string
output_type: string
output_type_id: string
value: double
model_id: string
reference_date: stringref_date_parquet_con <- arrow::open_dataset(ref_date_parquet_hubpath)
ref_date_parquet_conFileSystemDataset with 19 Parquet files
target: string
horizon: int32
target_end_date: date32[day]
location: string
output_type: string
output_type_id: string
value: double
model_id: string
reference_date: stringTarget hubs
target_csv_con <- arrow::open_dataset(target_csv_hubpath, format = "csv")
target_csv_conFileSystemDataset with 2 csv files
reference_date: date32[day]
horizon: int64
target_end_date: date32[day]
location: string
output_type: string
output_type_id: string
value: double
model_id: string
target: stringtarget_parquet_con <- arrow::open_dataset(target_parquet_hubpath)
target_parquet_conFileSystemDataset with 2 Parquet files
reference_date: date32[day]
horizon: int32
target_end_date: date32[day]
location: string
output_type: string
output_type_id: string
value: double
model_id: string
target: stringConnect to DuckDB model-output table
duckdb_con <- dbConnect(duckdb(), dbdir = duckdb_hub_path, read_only = TRUE)
# on.exit(dbDisconnect(duckdb_con))
model_out_db <- tbl(duckdb_con, "model_output")
model_out_db# Source: table<model_output> [?? x 9]
# Database: DuckDB v0.9.2 [Anna@Darwin 23.1.0:R 4.3.2/repartioned_hubs/flusight-hub.duckdb]
reference_date target horizon target_end_date location output_type
<date> <chr> <int> <date> <chr> <chr>
1 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
2 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
3 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
4 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
5 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
6 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
7 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
8 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
9 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
10 2023-10-14 wk inc flu hosp -1 2023-10-07 06 quantile
# ℹ more rows
# ℹ 3 more variables: output_type_id <chr>, value <dbl>, model_id <chr>Querying hubs for model ouput data for the latest n reference dates
I focus on a typical query for collecting data destined for downstream ensembling and visualization. The query is to get the latest n reference dates for a given target type.
Internal functions
I create a couple of internal functions for getting the latest n reference dates:
latest_n_ref_dates- Returns the latest n reference as a one column tibble.latest_n_ref_dates_vec- Returns the latest n reference as a vector.
latest_n_ref_dates <- function(hub_con, n = 6L) {
arrange(hub_con, desc(reference_date)) |>
distinct(reference_date) |>
head(n) |>
collect()
}
latest_n_ref_dates_vec <- function(hub_con, n = 6L) {
arrange(hub_con, desc(reference_date)) |>
distinct(reference_date) |>
head(n) |>
collect() |>
pull()
}Query functions
The two functions that actually perform the query via two different strategies are:
get_latest_inner_joinGets the latest n reference dates via inner joinget_latest_filterGets a vector of the latest n reference dates first and then retunrs hub data via filtering for ref dates
Functions also allow for additional filtering for target type or for returning data for all target types
get_latest_inner_join <- function(hub_con, target_type = "wk inc flu hosp", n = 6L) {
if (target_type == "all") {
# Only filter for ref dates
return(
inner_join(
hub_con,
latest_n_ref_dates(hub_con, n = n),
by = "reference_date"
) |>
collect()
)
}
# Filter for ref dates and target type
inner_join(
filter(hub_con, target %in% target_type),
latest_n_ref_dates(hub_con, n = n),
by = "reference_date"
) |>
collect()
}
get_latest_filter <- function(hub_con, target_type = "wk inc flu hosp", n = 6L) {
ref_dates <- latest_n_ref_dates_vec(hub_con, n = n)
if (target_type == "all") {
# Only filter for ref dates
return(
filter(
hub_con,
reference_date %in% ref_dates
) |>
collect()
)
}
# Filter for ref dates and target type
filter(
hub_con,
reference_date %in% ref_dates,
target %in% target_type
) |>
collect()
}Benchmarks
Vary number of rounds
For our benchmarks, we vary the number of n (number of latest rounds requested) from 1 to 18.
We also vary whether we are querying for all target types or for a single target type.
bp <- bench::press(
n = c(1L, 3L, 6L, 10L, 18L),
target_type = c("wk inc flu hosp", "all"),
{
bench::mark(
std_hub_csv_filter = get_latest_filter(baseline_csv_hub_con,
n = n,
target_type = target_type
),
std_hub_csv_inner_join = get_latest_inner_join(baseline_csv_hub_con,
n = n,
target_type = target_type
),
std_hub_parquet_filter = get_latest_filter(baseline_parquet_hub_con,
n = n,
target_type = target_type
),
std_hub_parquet_inner_join = get_latest_inner_join(baseline_parquet_hub_con,
n = n,
target_type = target_type
),
target_parquet_filter = get_latest_filter(target_parquet_con,
n = n,
target_type = target_type
),
ref_date_parquet_filter = get_latest_filter(ref_date_parquet_con,
n = n,
target_type = target_type
),
target_parquet_inner_join = get_latest_inner_join(target_parquet_con,
n = n,
target_type = target_type
),
ref_date_parquet_inner_join = get_latest_inner_join(ref_date_parquet_con,
n = n,
target_type = target_type
),
target_csv_filter = get_latest_filter(target_csv_con,
n = n,
target_type = target_type
),
ref_date_csv_filter = get_latest_filter(ref_date_csv_con,
n = n,
target_type = target_type
),
target_csv_inner_join = get_latest_inner_join(target_csv_con,
n = n,
target_type = target_type
),
ref_date_csv_inner_join = get_latest_inner_join(ref_date_csv_con,
n = n,
target_type = target_type
),
duckDB = get_latest_filter(model_out_db,
n = n,
target_type = target_type
),
check = FALSE
)
}
)Running with:
n target_type 1 1 wk inc flu hosp 2 3 wk inc flu hospWarning: Some expressions had a GC in every iteration; so filtering is
disabled. 3 6 wk inc flu hospWarning: Some expressions had a GC in every iteration; so filtering is
disabled. 4 10 wk inc flu hospWarning: Some expressions had a GC in every iteration; so filtering is
disabled. 5 18 wk inc flu hospWarning: Some expressions had a GC in every iteration; so filtering is
disabled. 6 1 all Warning: Some expressions had a GC in every iteration; so filtering is
disabled. 7 3 all Warning: Some expressions had a GC in every iteration; so filtering is
disabled. 8 6 all Warning: Some expressions had a GC in every iteration; so filtering is
disabled. 9 10 all Warning: Some expressions had a GC in every iteration; so filtering is
disabled.10 18 all Warning: Some expressions had a GC in every iteration; so filtering is
disabled.plot(bp)Loading required namespace: tidyr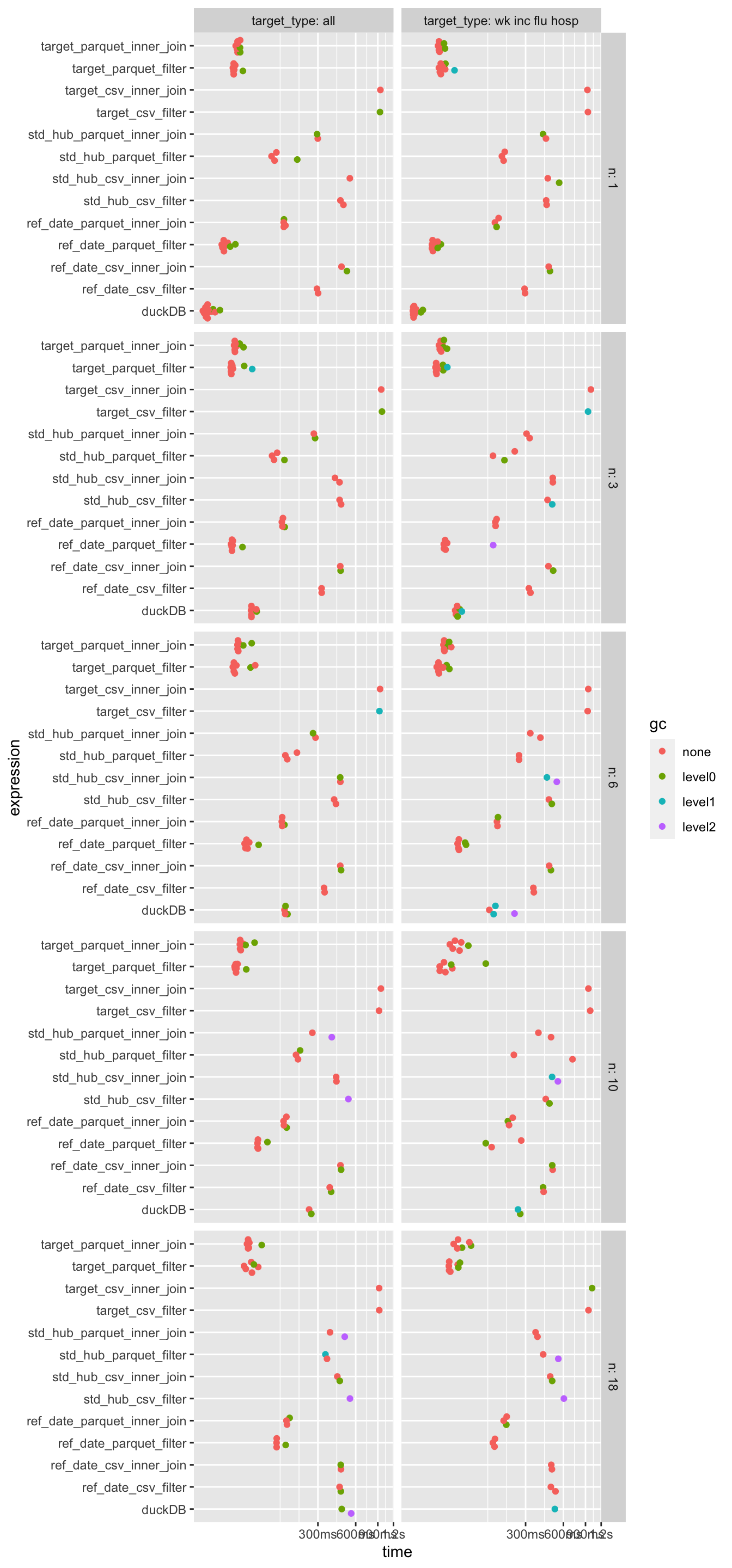
APPENDIX - Creating re-partitioned hubs
The basis for the re-partitioned hub is the current FluSight hub. The hub is available at: https://github.com/cdcepi/FluSight-forecast-hub
The Flusight hub was cloned and placed in the same parent directory as this benchmarking repository (i.e. is a sibling directory to this repository).
The hub was cloned and commit 6df410 was checked out to create a new branch “temp” containing all git history up to that commit.
The hub was then re-partitioned by reference_date or target and saved as CSV or Parquet formatted hub.
The hub was also re-partitioned by model_id and reference_date and saved as Parquet to compare a parquet version of a std hub.
The standard hub was also copied to the re-partitioned hub directory.
Finally, the hub was also converted to a DuckDB database file.
library(arrow)
library(hubUtils)
library(dplyr)
# Create a new branch ("temp") containing all git history up to a specific commit ("6df410").
commit <- git4r::git_get(hash = "6df410")
git4r::git_checkout(
commit = commit,
onto_new_branch = "temp"
)
# Checkout "temp" branch before connecting to std hub and collecting all data
hub_con <- connect_hub(hub_path = ".")
hub_data <- hub_con |> collect()
# Function to repartition hub
repartition_hub <- function(partition = "reference_date", format = "csv",
repartitioned_hub_dir = here::here(
"../partition-benchmarks/repartioned_hubs"
)) {
fs::dir_create(repartitioned_hub_dir)
hub_name <- paste("flusight-hub", paste(partition, collapse = "-"), format, sep = "_")
part_hub_path <- fs::path(repartitioned_hub_dir, hub_name, "model-output")
part_hub_config_path <- fs::path(repartitioned_hub_dir, hub_name, "hub-config")
fs::dir_create(part_hub_config_path)
write_dataset(hub_data,
format = format,
path = part_hub_path,
partitioning = partition
)
fs::dir_copy("hub-config", part_hub_config_path, overwrite = TRUE)
}
# SET PATH TO REPARTITIONED HUB DIRECTORY
repartitioned_hub_dir <- here::here("../partition-benchmarks/repartioned_hubs")
# Create repartitioned hubs
# Partitions by reference_date - csv
repartition_hub()
# Partitions by target - csv
repartition_hub("target")
# Partitions by reference_date - parquet
repartition_hub(format = "parquet")
# Partitions by target - parquet
repartition_hub("target", format = "parquet")
# Partitions by model_id - parquet
repartition_hub("model_id", format = "parquet")
# Partitions by model_id & reference_date - parquet - reflects std hub partitioning
# but as parquet
repartition_hub(c("model_id", "reference_date"), format = "parquet")
# Copy std hub
fs::dir_copy("hub-config", fs::path(repartitioned_hub_dir, "flusight-hub_std_csv", "hub-config"), overwrite = TRUE)
fs::dir_copy("model-output", fs::path(repartitioned_hub_dir, "flusight-hub_std_csv", "model-output"), overwrite = TRUE)
library("duckdb")
# to use a database file (not shared between processes)
con <- dbConnect(duckdb(), dbdir = fs::path(repartitioned_hub_dir, "flusight-hub.duckdb"), read_only = FALSE)
dbWriteTable(con, "model_output", hub_data, overwrite = TRUE)
dbDisconnect(con, shutdown = TRUE)Session Info
devtools::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os macOS Sonoma 14.1.2
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Athens
date 2024-02-14
pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
arrow 14.0.0.2 2023-12-02 [1] CRAN (R 4.3.1)
askpass 1.2.0 2023-09-03 [1] CRAN (R 4.3.0)
assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.3.0)
backports 1.4.1 2021-12-13 [1] CRAN (R 4.3.0)
beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.3.0)
bench 1.1.3 2023-05-04 [1] CRAN (R 4.3.0)
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.0)
brio 1.1.4 2023-12-10 [1] CRAN (R 4.3.1)
bslib 0.6.1 2023-11-28 [1] CRAN (R 4.3.1)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.0)
checkmate 2.3.1 2023-12-04 [1] CRAN (R 4.3.1)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.1)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
credentials 2.0.1 2023-09-06 [1] CRAN (R 4.3.0)
crosstalk 1.2.1 2023-11-23 [1] CRAN (R 4.3.1)
DBI * 1.2.0 2023-12-21 [1] CRAN (R 4.3.1)
dbplyr 2.4.0 2023-10-26 [1] CRAN (R 4.3.1)
devtools * 2.4.5 2022-10-11 [1] CRAN (R 4.3.0)
digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
DT 0.31 2023-12-09 [1] CRAN (R 4.3.1)
duckdb * 0.9.2-1 2023-11-28 [1] CRAN (R 4.3.1)
ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.3.0)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
fs 1.6.3 2023-07-20 [1] CRAN (R 4.3.0)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
ggbeeswarm 0.7.2 2023-04-29 [1] CRAN (R 4.3.0)
ggplot2 3.4.4 2023-10-12 [1] CRAN (R 4.3.1)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
httpuv 1.6.14 2024-01-26 [1] CRAN (R 4.3.1)
hubUtils * 0.0.0.9018 2024-01-30 [1] local
jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
later 1.3.2 2023-12-06 [1] CRAN (R 4.3.1)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.0)
mime 0.12 2021-09-28 [1] CRAN (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
openssl 2.1.1 2023-09-25 [1] CRAN (R 4.3.1)
pak * 0.7.1 2023-12-11 [1] CRAN (R 4.3.1)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgbuild 1.4.3 2023-12-10 [1] CRAN (R 4.3.1)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
pkgload 1.3.3 2023-09-22 [1] CRAN (R 4.3.1)
profmem 0.6.0 2020-12-13 [1] CRAN (R 4.3.0)
profvis 0.3.8 2023-05-02 [1] CRAN (R 4.3.0)
promises 1.2.1 2023-08-10 [1] CRAN (R 4.3.0)
purrr 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.1)
remotes 2.4.2.1 2023-07-18 [1] CRAN (R 4.3.0)
reprex * 2.0.2 2022-08-17 [1] CRAN (R 4.3.0)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.1)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
sass 0.4.8 2023-12-06 [1] CRAN (R 4.3.1)
scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.1)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
shiny 1.8.0 2023-11-17 [1] CRAN (R 4.3.1)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
stringr 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
sys 3.4.2 2023-05-23 [1] CRAN (R 4.3.0)
testthat * 3.2.1 2023-12-02 [1] CRAN (R 4.3.1)
tibble 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidyr 1.3.1 2024-01-24 [1] CRAN (R 4.3.1)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
urlchecker 1.0.1 2021-11-30 [1] CRAN (R 4.3.0)
usethis * 2.2.2 2023-07-06 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vipor 0.4.7 2023-12-18 [1] CRAN (R 4.3.1)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.1)
xfun 0.41 2023-11-01 [1] CRAN (R 4.3.1)
xtable 1.8-4 2019-04-21 [1] CRAN (R 4.3.0)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
[1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────